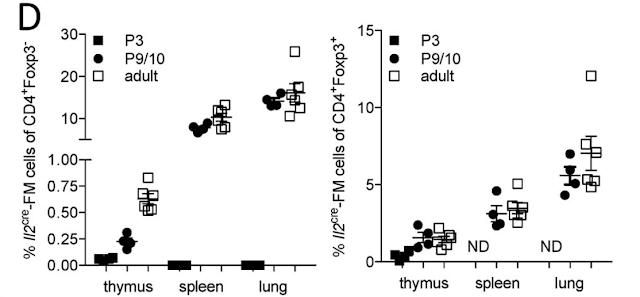
So far 3 different outcomes have been identified for developing T cells in the thymus: to develop into naive T cells, get deleted or become Foxp3+ Treg. Both deletion and Treg path require the presence of specific epitopes. However, how a given T cell decides between these pathways is not well understood.
Here is a new paper in PNAS that tries to tackle this question using the tetramer tracking approach. The authors are using PLP (brain-specific protein) as an endogenous antigen expressed in the thymus. Surprisingly both PLPWT and PLPKO mice showed near similar numbers of tetramer-positive T cells in peripheral tissue. However, as expected, only PLPWT mice that express PLP epitopes in the thymus but not PLPKO mice that do not express the same epitopes showed Treg development.
Similar results were obtained when thymus tissue was analyzed.
To make tetramer tracking for reliable the authors used transgenic mice expressing a fixed TCR beta chain. These mice also showed a similar phenotype.
As in PLPWT and PLPKO mice, fixed:TCR beta mice on PLPWT but not on PLPKO background harbored Tregs in the periphery. Notable, the rest of the tetramer-positive Foxp3-negative T cells displayed an anergic phenotype (CD73HiFR4Hi).
A similar phenotype was found in the thymus. Note, there was an unexpected and significant reduction of tetramer-positive T cells from the thymus to the periphery in fixed:TCR beta mice on PLPKO background compared to fixed:TCR beta mice on PLPWT background.
So far these data indicated that there is almost no deletion of PLP specific T cells in the thymus on WT mice [compaed KO] but ~2-fold reduction in fixed:TCR beta mice on PLPWT compared to KO. Almost half of the tetramer-positive T cells ended up in the Treg pool on the WT background. The remaining T cells showed an anergic phenotype. However the dramatic reduction of tetramer-positive T cells from the thymus to the periphery in KO mice raises some serious unanswered questions.
Finally, to find some correlation between TCR specificity and Treg/anergy/deletion phenotype, the authors selected 4 PLP-specific TCRs (denoted here as A, B, C, D). Their analysis showed that some (clone "A") but not other PLP-specific TCRs (clone "C") were able to generate Tregs in the thymus. Notable, TCR "C" displayed the highest affinity to PLP epitope. Also, there is a substantial reduction of clone "C" from the thymus to the periphery in the Foxp3-negative compartment. This possibly reflects the fact that most clones in "C" are anergic and slowly disappear from the periphery.
In summary, this study re-confirms that tolerance to self-antigens is mostly controlled via Treg generation and that not all antigens/epitopes and their corresponding TCRs are able to participate in this process. There are few unexplained observations in this paper though as discussed above.
posted by David Usharauli